Речевая аналитика — обеспечивает конвертацию разговоров в текстовый формат, что открывает дополнительные возможности для
На примере сервиса Ucaller, предоставляющего такой сервис, можно увидеть, что доставлять коды можно двумя
Profit Server уже почти 10 лет работает в сфере предоставления услуг по аренде выделенных
Ищете хостинг или виртуальный сервер с комфортными условиями по оплате и использованию? Такие решения
Перечень операций в рамках настройки 1С и стоимость услуг зависит от специфики компании, внедряемого/настраиваемого
При заказе VPS/VDS от CloudVPS можно воспользоваться готовыми дистрибутивами для быстрого старта.
Эквайринг – это процесс обработки платежей с использованием банковских карт, который стал неотъемлемой частью
С развитием технологий и расширением возможностей онлайн-связи, это становится все более привлекательным и востребованным
JUSTHOST — провайдер услуг, у которого есть все необходимое для уверенного присутствия в интернете.
Согласно статистике ежегодно российские маркетплейсы растут x2. Это касается, как заработка самих площадок, так
Regfon, как комплексное решение, включает услуги по продаже/аренде виртуальных номеров, ВАТС и оборудования для
Виртуальный номер телефона США (1+) – это удобный и эффективный способ общения и ведения
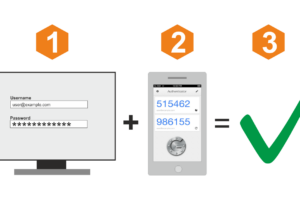
Ниже — обзор популярных сервисов авторизации по звонку, с помощью которых вы сможете быстро
Теплые полы – это современная система обогрева, которая обеспечивает равномерное распределение тепла по поверхности
Зимой уборка крыш от снега альпинистами очень необходима для безопасности и комфорта жителей города.
Системный интегратор информационно-коммуникационных технологий (ИКТ) является ключевым звеном в цепочке создания эффективных и гармонично
С течением времени технологии стремительно развиваются, и то, что казалось современным и актуальным несколько
Различные типы ковров требуют разных подходов к стирке. Например, натуральные шерстяные ковры требуют более
Полиграфия является важной составляющей современного мира коммуникаций.
Со временем ванные комнаты могут подвергаться износу и старению. Однако, перед тем как считать,
Венки можно встретить на многих захоронениях. Выбирают их в дань памяти ушедшему, показывая свое
Выбор виртуального номера телефона может быть сложной задачей, но существует несколько факторов, которые следует
Веб-шрифты – это особые шрифты, которые загружаются и отображаются на веб-страницах, позволяя веб-дизайнерам использовать
Восстановление секторов жесткого диска - это процесс восстановления поврежденных или неисправных секторов на жестком
В современном мире, где коммуникация играет ключевую роль в развитии бизнеса, виртуальные номера телефонов
Регистрация медицинских изделий является важной процедурой, предшествующей их введению на рынок и использованию в
Смартфоны стали неотъемлемой частью нашей повседневной жизни. Мы используем их для общения, развлечений и
Производство холодильного оборудования – пошаговая инструкция по организации бизнеса. Анализ рынка + регистрация ИП
Интерьерная печать – это уникальный и эффективный способ преобразить обычное пространство в неповторимое и